Motivation
Genetic mutations in specific genes elevate the risk of developing genetic diseases, collectively called hereditary genetic syndromes. Understanding associations of genetic mutations with these syndromes helps tailor diagnostic and preventive measures for carriers of mutations.
Classification of variants identified by a typical sequencing or panel assay requires one or more types of evidence. At least eight evidence codes (PP1, PM6, PS2, PS3, PP4, BS3, BS4, and BP3) described in ACMG guidelines use case reports, case series and assay results in one form or another to support variant classification.
Expert-led curation efforts help collect, curate, and compile data for variation classification. However, such efforts are expensive, and the scale needs to be improved. We have identified essential components and automated relevant evidence collection and curation processes without human intervention.
The Adenine AI (Clinical Evidence For Genomic Medicine) is an output of a fully automated data curation pipeline. The pipeline collects, curates, and normalizes data for knowledge discovery. The Adenine AI comprises knowledge across three domains: case reports, case series, and assays. Case reports include a detailed description of one patient, and case series are outcomes of studies on multiple patients. Assays help to understand the impact of mutations in model systems.

Contents
Our platform strives to integrate the latest discoveries and insights as they become available. However, it's important to note that our ability to update content is limited by the availability of abstracts from publications, some of which may not be available in PubMed, and we do not currently have access to full texts or supplementary materials.
Adenine AI's approach to supporting the nuanced needs of genetic variant classification and gene-disease association is grounded in use of AI to identify and organize evidence. While being comprehensive is not at all the objective of this resource, we try to provide more information per click. We categorizing evidence into three distinct categories—case reports, multi-patient studies, and functional assays—Adenine AI aims to address specific aspects under the ACMG/AMP guidelines.
Case Reports:
Case reports offer granular insights into the phenotypic expression and clinical impact of genetic variants in individual patients. They enrich the database with structured curation that can uncover novel associations and rare variant manifestations.
Multi-patients studies:
Multi-patient studies provide a robust framework for understanding genetic variants across larger populations, offering statistical insights into variant frequency, penetrance, and disease association. This collective data is crucial for establishing the clinical significance of genetic alterations.
Functional assays:
Functional assays directly measure the biological effects of genetic variants on gene or protein function, offering essential clues about the pathogenicity of variants of uncertain significance (VUS). These assays are key to linking genetic changes with their phenotypic outcomes, guiding therapeutic decision-making.
The interface
Adenine AI’s interface is crafted with user experience at the forefront, ensuring that navigation is fast, straightforward, and effective. The design principles focus on minimizing the learning curve, allowing users to quickly become proficient in using the platform. This intuitive nature means usable evidence is just a few clicks away.
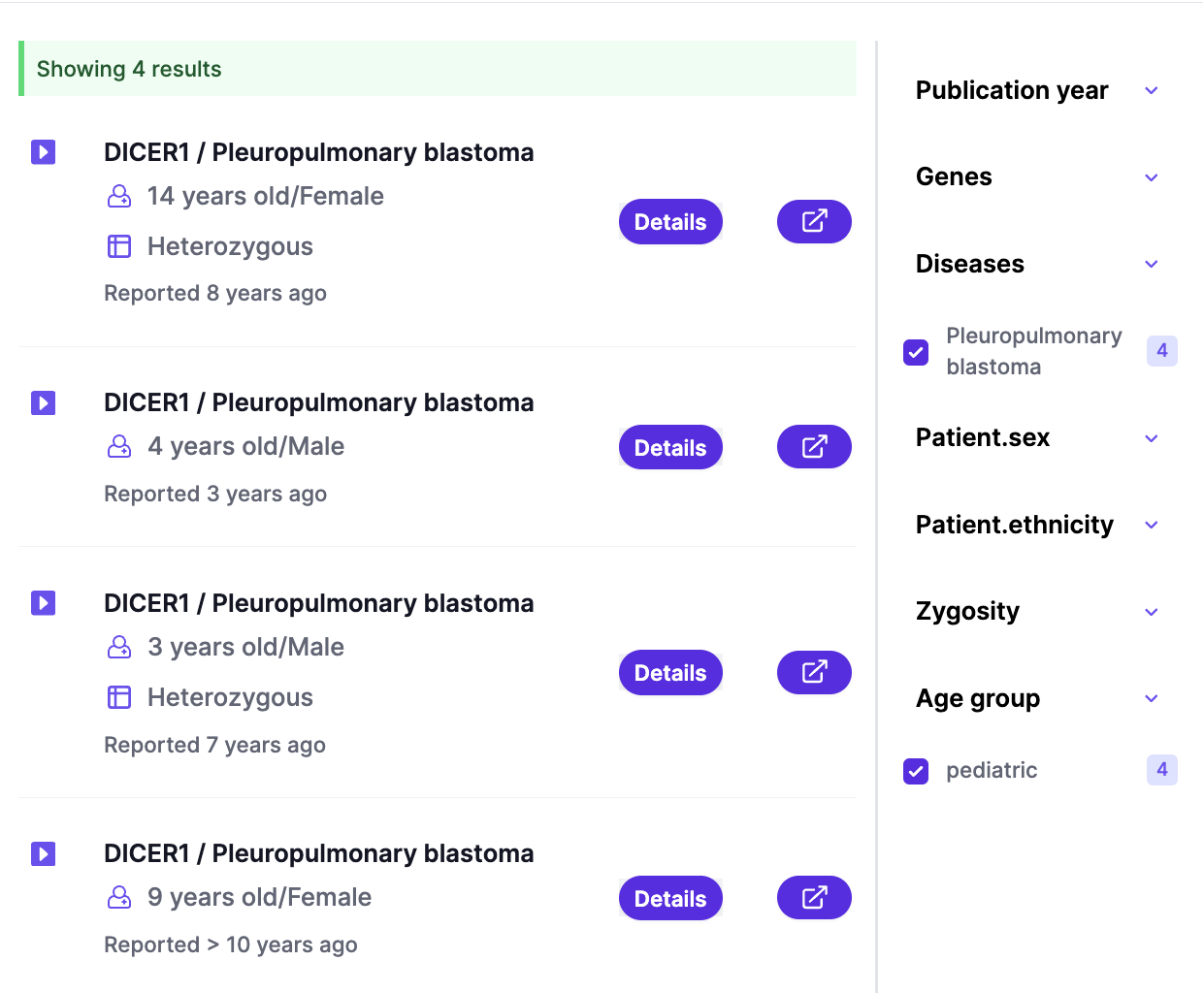
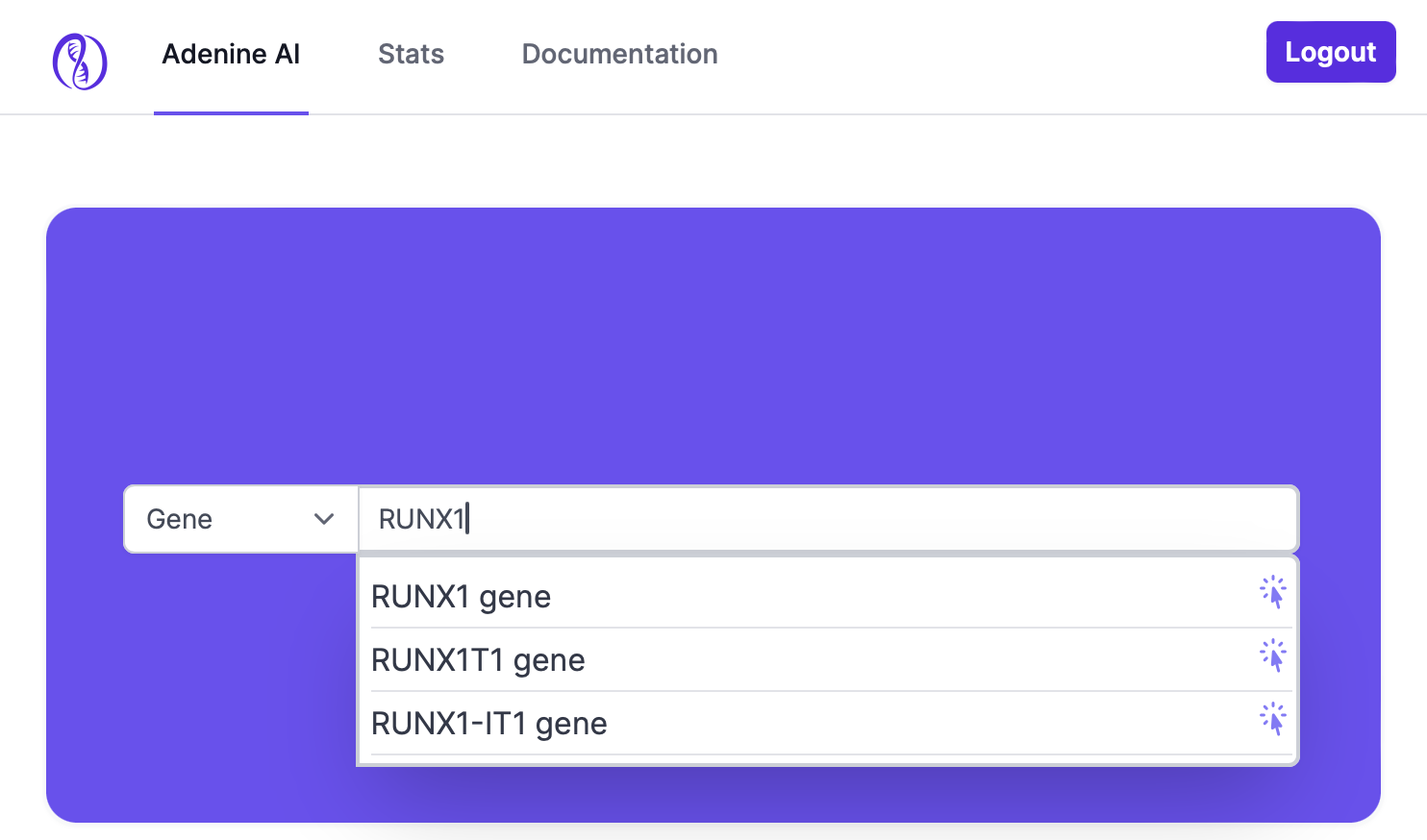
Adenine AI simplifies the evidence discovery process by enabling a single search query to span across all evidence categories: case reports, multi-patient studies, and functional assays. This unified search approach ensures that users can easily access a holistic view of the genetic variant under investigation without having to perform multiple searches.
An initial search with gene, variant, or phenotype combined with powerful facets and filter options helps refine search results to reach to evidence of interest. Users can narrow down their search results based on various parameters, such as patient characteristics (e.g., age, sex, ethnicity), cohort composition (e.g., disease prevalence, control groups), and other critical factors. These facets allow for the customization of search queries to match specific needs, ensuring that the evidence presented is as pertinent and actionable as possible.
By focusing on an intuitive interface, powerful search capabilities, a unified search feature, and advanced filtering options, Adenine AI ensures that users have a seamless and productive experience. These features underscore the platform’s commitment to supporting the genetic counseling and molecular pathology communities with a tool that is not only advanced but also user-friendly and highly adaptable to their needs.